Current Focus: Decoding Nuclear Biocondensates in 3D
I investigate how spatial organization of nuclear condensates (Cajal bodies, PML-nuclear bodies, speckles, nucleoli) correlates with cellular function and aging. My work combines:
- Advanced microscopy (confocal, super-resolution)
- Custom computational pipelines for 3D segmentation/analysis
- Open-source tool development to democratize spatial cytology
Why 3D Matters
While confocal microscopy generates rich 3D datasets, 2D projections require fewer computational resources and are easier to interpret (MDPI, 2021). This discards critical spatial information about:
- Liquid-liquid phase separation (LLPS) dynamics
- Chromatin-nuclear body interactions
- Anisotropic distribution patterns linked to gene activity
My tools extract this “hidden” data to reveal how nuclear architecture encodes function. I aim to provide a simmpler aproach to bridge bioinformatics and cellular morphology.
Featured Work: Nuclear Analysis Suite
3Dnucleus_data
An open-source ImageJ/R pipeline for high-throughput analysis of nuclear condensates
To really understand how the tools we use in cytology work, I built my own image analysis pipeline using ImageJ macros. Writing the code myself forced me to learn exactly how these methods process and quantify cellular data — no magic boxes, just full control and transparency.
Key Features:
- Processes 1000+
.liffiles in <3h - Quantifies:
- Cajal/PML body volume, intensity, spatial distribution
- Chromatin proximity (DAPI overlap)
- LLPS-driven clustering patterns
- Modular design adaptable to other condensates (e.g., nucleoli, histone locus bodies)
Example Output:
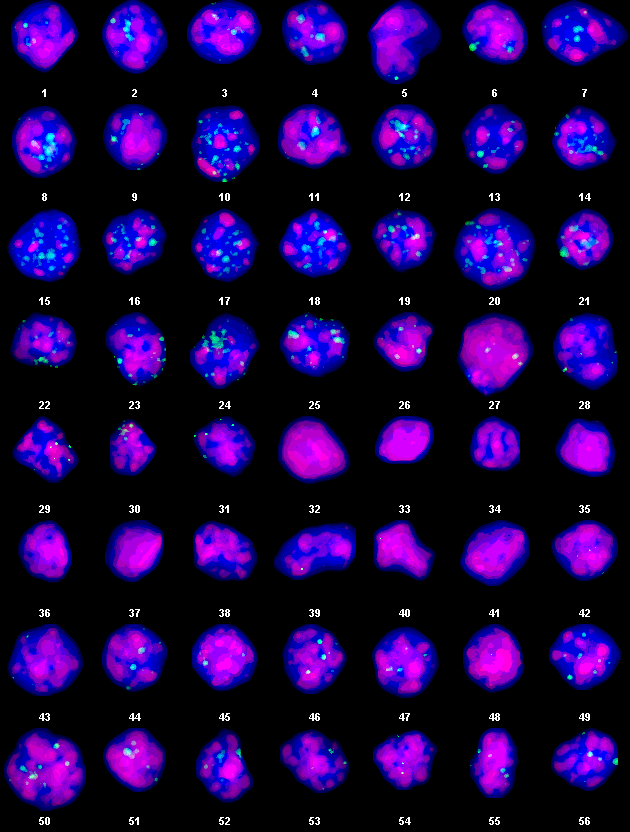
Automated segmentation of coilin+ bodies (green) concentrated DAPI regions (red) and poorly-stainded DAPI regions (Blue) in cumulus cell nuclei.
Example of macro workflow for statistical data extraction:
Volume distribution (kernel density estimate) of individual coilin+ body volumes within each nucleus (n=12). Violin graphs, Rstudio
Recent Work: Educational 3D nucleus reconstructon
As part of my research in cellular imaging, I successfully generated a 3D volumetric reconstruction of a cumulus cell nucleus using serial confocal microscopy images. This project combined advanced imaging techniques with computational 3D modeling to create an interactive educational tool for visualizing nuclear morphology.
3D reconstruction of segmented nucleus objects from a cumulus cell (located in mammalian follicle). Segmented objects – nuclear envelope, DAPI regions and coilin+ bodies.

